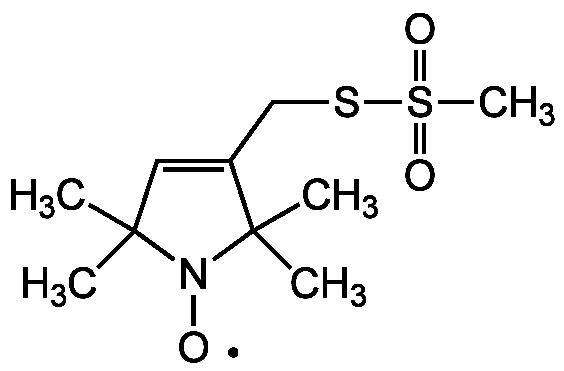
Chemical Structure
MTSSL [81213-52-7]

AG-CR1-3573
CAS Number81213-52-7
Product group Chemicals
Estimated Purity>98%
Molecular Weight264.3
Overview
- SupplierAdipoGen Life Sciences
- Product NameMTSSL [81213-52-7]
- Delivery Days Customer10
- CAS Number81213-52-7
- CertificationResearch Use Only
- Estimated Purity>98%
- Hazard InformationNon-hazardous
- Molecular FormulaC10H18NO3S2
- Molecular Weight264.3
- Scientific DescriptionChemical. CAS: 81213-52-7. Formula: C10H18NO3S2. MW: 264.3. Highly reactive, thiol-specific spin-label. Specific conformational probe of thiol site structure by its minimal rotational freedom and distance from the covalent disulfide linkage to the macromolecule under study. Used to label cysteine residues in proteins (site-directed labeling, SDS-labeling). Allows protein structure and protein dynamics determination as well as the study of protein-protein and protein-oligonucleotide interactions. - Highly reactive, thiol-specific spin-label. Specific conformational probe of thiol site structure by its minimal rotational freedom and distance from the covalent disulfide linkage to the macromolecule under study. Used to label cysteine residues in proteins (site-directed labeling, SDS-labeling). Allows protein structure and protein dynamics determination as well as the study of protein-protein and protein-oligonucleotide interactions.
- SMILESCC1(C)C=C(CSS(C)(=O)=O)C(C)(C)N1[O]
- Storage Instruction2°C to 8°C,-20°C
- UNSPSC12352200
References
- A novel reversible thiol-specific spin label: papain active site labeling and inhibition: L.J. Berliner, et al.; Anal. Biochem. 119, 450 (1982)
- ESR spectra reflect local and global mobility in a short spin-labeled peptide throughout the alpha-helix-coil transition: A.P. Todd & G.L. Millhauser; Biochemistry 30, 5515 (1991)
- Kinetics and motional dynamics of spin-labeled yeast iso-1-cytochrome c: 1. Stopped-flow electron paramagnetic resonance as a probe for protein folding/unfolding of the C-terminal helix spin-labeled at cysteine 102: K. Qu, et al.; Biochemistry 36, 2884 (1997)
- Pressure-induced thermostabilization of glutamate dehydrogenase from the hyperthermophile Pyrococcus furiosus: M.M. Sun, et al.; Protein Sci. 8, 1056 (1999)
- Protein global fold determination using site-directed spin and isotope labeling: V. Gaponenko, et al.; Protein Sci. 9, 302 (2000)
- High-resolution probing of local conformational changes in proteins by the use of multiple labeling: unfolding and self-assembly of human carbonic anhydrase II monitored by spin, fluorescent, and chemical reactivity probes: P. Hammarstroem, et al.; Biophys. J. 80, 2867 (2001)
- Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme: P.P. Borbat, et al.; JACS 124, 5304 (2002)
- Methods for study of protein dynamics and protein-protein interaction in protein-ubiquitination by electron paramagnetic resonance spectroscopy: H.J. Steinhoff; Front. Biosci. 7, c97 (2002)
- Inter- and intra-molecular distances determined by EPR spectroscopy and site-directed spin labeling reveal protein-protein and protein-oligonucleotide interaction: H.J. Steinhoff; Biol. Chem. 385, 913 (2004)
- Dynamics of the nitroxide side chain in spin-labeled proteins: F. Tombolato, et al.; J. Phys. Chem. B. 110, 26248 (2006)
