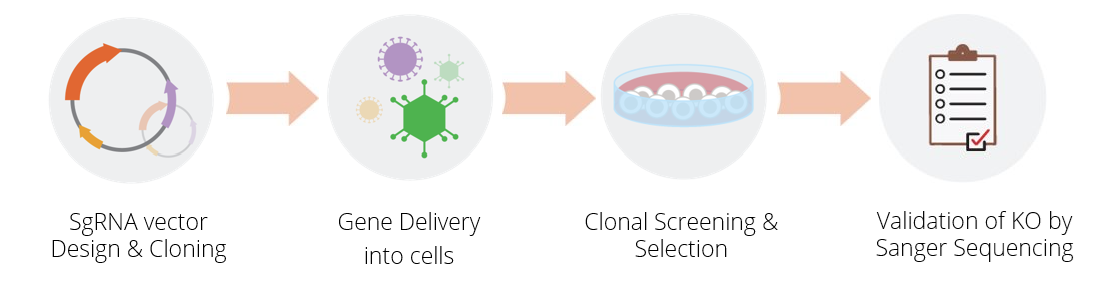
Bio-Connect Joins the European Distributor Meeting of abm
Bio-Connect welcomes its new supplier, abm, which provides advanced cell biology solutions, including cell lines, CRISPR gene editing, and molecular biology tools.
abm’s CRISPR Knockout Cell Line Generation Service offers a turnkey solution—handling everything from design to validation—so researchers can accelerate discoveries without the burden of cell line development.
The CRISPR-Cas9 system has become a cornerstone in modern gene editing, allowing researchers to unravel gene function with unprecedented precision. abm’s CRISPR Knockout (KO) Cell Line Service offers a complete, hassle-free solution to generate stable gene knockout cell lines—empowering functional studies, disease modeling, and drug screening with reliable, ready-to-use models.
abm also offers a wide selection of pre-made knockout cell lines targeting key genes across various biological pathways—shipped ready to use, with full validation data.
We are quite happy with the knockout cells that you made. This was a tricky knockout and hemizygous removal was much more prevalent than the double knockout, but you did it. In addition to your excellent genomic sequence characterization of both mutant LIF alleles, I have confirmed in our lab that the tumor cells have lost all mouse LIF production.”
– Dr. Robert Jackman, Boston University, Mouse LIF CRISPR Knock Out C26 Tumor

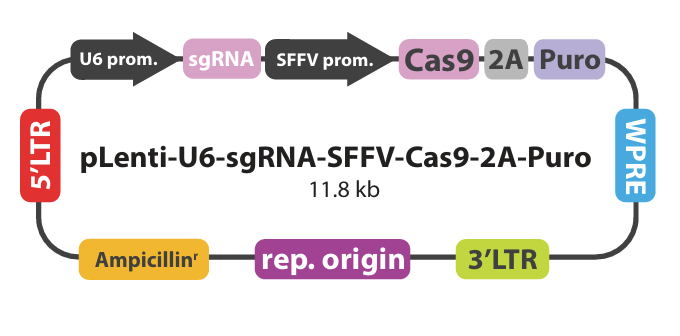
The LIF (Leukemia Inhibitory Factor) gene, implicated in tumor progression and cachexia, was targeted using abm’s lentiviral CRISPR/Cas9 system. Through a rigorous multi-phase workflow—spanning sgRNA design, lentiviral transduction, puromycin selection, colony screening, and next-generation sequencing—abm isolated a monoclonal cell line (Clone 6a) with confirmed biallelic frameshift mutations at the genetic level and complete knockout at the protein level.
Three sgRNA were designed against mouse LIF locus (Mus musculus, NM_008501). Software analysis was performed to ensure the sgRNA had no predicted off-targets binding sites.
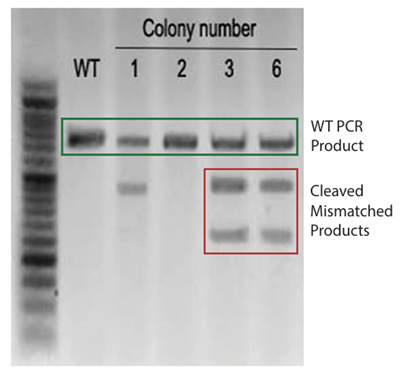
Cell colonies are isolated after puromycin selection. Genomic DNA was extracted and the surveyor assay was performed to confirm genomic editing of the LIF locus.
Figure 2: The surveyor assay indicated that Colony 3 and 6 were edited; colony 2 was not edited; and colony 1 was inconclusive.
PCR products from Colonies 3 and 6 were further analyzed via Sanger Sequencing to determine the nature of the knockout (Figure 3). For colony 3 only one mutant sequence was detected, indicating that these cells are likely only heterozygotic knockouts. In colony 6 two different mutant sequences were detected.

Colony 6 was serial diluted into 96 well plates for monoclonal selection. Genomic DNA was extracted from these clones (i.e. 6a, 6b..), PCR amplified, cloned and sequenced.
Of the colony 6 clones, sequencing showed that only clone 6a had a frameshift mutation in both alleles (Figure 4). A frameshift mutation disrupts the open reading frame, resulting in nonsense mediated decay of mRNA transcript. Further sequencing of clone 6a confirmed that only two mutant alleles were present, the 2 bp and 4 bp deletions, and that no WT or other mutations were detected (Figure 5).
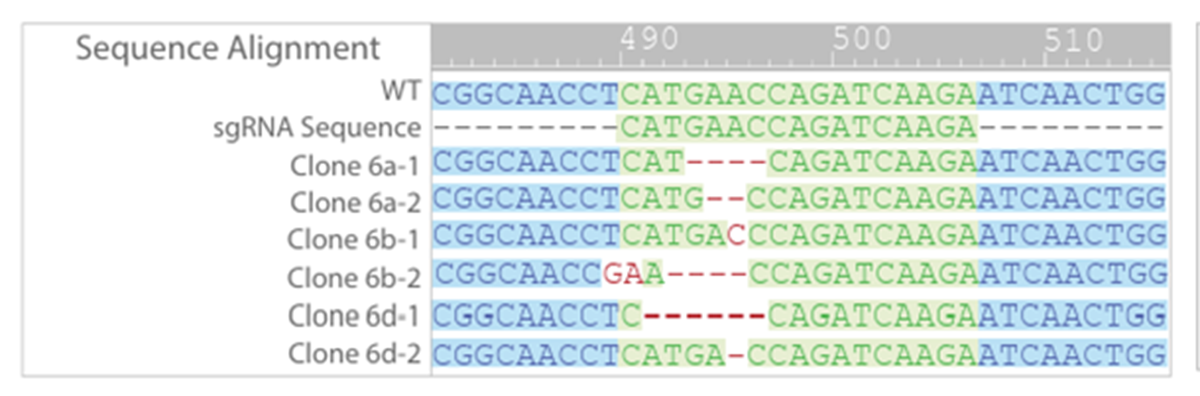
Figure 4: Clones 6a, 6b and 6d all showed biallelic editing. Only clone 6a had frame shift mutations in both alleles. No WT sequences were detected in all subclones.

Figure 5: Further sequencing of 6a confirmedn bialleelic knock-out. No WT sequences were detected.
Next generation sequencing was performed at each stage of selection to evaluate knockout (Figure 6). Before editing, only WT sequences were observed. After the first round of selection colony 6 showed a mixture of edited (70%) and WT (30%) sequences. Finally, after monoclonal selection, clone 6a showed only edited sequences with no WT alleles present
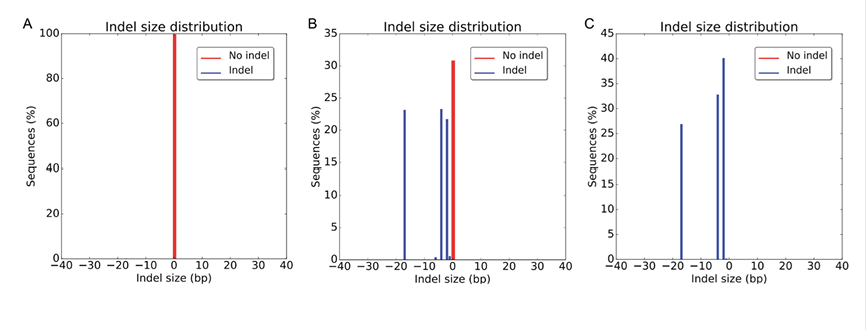

Figure 7: LIF Secretion into media by the resulting KO cell line was tested via ELISA.The wild-type C26 cells secreted substantial levels of LIF while C26 LIF KO cell line secreted undetectable or no LIF.Data from Kandarian et al., 2018. J Cachexia Sarcopenia Muscle, 9(5), 986–998.
Tumour-derived leukaemia inhibitory factor is a major driver of cancer cachexia and morbidity in C26 tumour-bearing mice. Read more: https://doi.org/10.1002/jcsm.12346
We gladly support you by keeping you updated on our latest products and the developments around our services.